Authors: Josep M. Badia [aut, cre] (https://orcid.org/0000-0002-5704-1124)
Last modified: 2021-12-03 16:51:03
Compiled: Fri Jan 7 20:40:30 2022
Introduction
MS2ID aims to fast annotate query MS/MS spectra straightforwardly with minimum RAM requirements by using an in-house database. With this in mind, MS2ID object keeps all data in a on disk backend based on RSQLite and bigmemory packages and indexes all spectrum fragments in a table.
Conceptually, MS2ID workflow is structured on three steps:
-
Creating an in-house MSn spectra library as a
M2IDobject- MS2IDdirectory <- createMS2ID(Mona, …)
-
Annotating MSn query spectra against an MS2ID object, obtaining as a result an
Annotobject- MS2IDobject <- MS2ID(MS2IDdirectory)
- AnnotObject <- annotate(querySpectra, MS2IDobject, …)
- MS2IDobject <- MS2ID(MS2IDdirectory)
-
Browsing the results using the MS2ID GUI interface based on Shiny
- MS2IDgui(AnnotObject)
Installation
You can download the development version from GitHub with:
if(!requireNamespace("devtools", quietly = TRUE))
install.packages("devtools")
devtools::install_github("jmbadia/MS2ID", force=T)MS2ID creation
MS2ID package already contains a sample MS/MS spectra library. Next code extracts it in order to be used:
## Decompress the MS2ID library that comes with MS2ID
MS2IDzipFile <- system.file("extdata/MS2IDLibrary.zip", package = "MS2ID",
mustWork = TRUE)
library(utils)
MS2IDdirectory <- dirname(utils::unzip(MS2IDzipFile, exdir = tempdir()))[1]On the other hand, reference MS/MS spectral data can be obtained from publicly available resources -HMDB, ChEBI and PubChem or any personal library- through the CompoundDb package (Stanstrup and Rainer 2021) plus the createMS2ID function. Here we parse a small portion of MoNA (206 MS/MS spectra) following the CompoundDB vignette.
library(CompoundDb)
wrkDir <- tempdir()
## Locate the compounds file
MoNAsubset <- system.file("extdata/MoNAsubset.sdf.gz", package = "MS2ID")
cmps <- compound_tbl_sdf(MoNAsubset)
spctr <- msms_spectra_mona(MoNAsubset, collapsed = TRUE)
spctr$predicted <- FALSE
#configure metadata
metad <- data.frame(
name = c("source", "url", "source_version","source_date"),
value = c("MoNA", "https://mona.fiehnlab.ucdavis.edu/downloads",
"v1", '07-09')
)
#obtain compdb object
cmpdbDir <- file.path(wrkDir, "cmpdbDir")
if(dir.exists(cmpdbDir)){
do.call(file.remove, list(list.files(cmpdbDir, full.names = TRUE)))
}else{
dir.create(cmpdbDir)
}
db_file <- createCompDb(cmps, metadata = metad, msms_spectra = spctr,
path = cmpdbDir)
cmpdb <- CompDb(db_file)
#create the MS2ID directory
library(MS2ID)
MS2IDdirectory <- createMS2ID(cmpdb = cmpdb,
overwrite = TRUE, path = wrkDir)Annotation
A simple annotation only requires an MS2ID object like the former and query spectra. In the example following, query spectra are provided in mzML format (three mzML files, 2235 MS/MS spectra total). Still, the function also supports Spectra objects from the Spectra package (Gatto, Rainer, and Gibb 2021).
By default, the annotation process summarizes query spectra into consensus spectra according to their similarity and neighbouring; this speeds up the algorithm and reduces the noise. Please, consult the annotation vignette for more information.
library(utils)
queryFile <- system.file("extdata/QRYspectra.zip", package = "MS2ID")
queryFolder <- file.path(tempdir(), "QRYspectra")
utils::unzip(queryFile, exdir = queryFolder)
refLibrary <- MS2ID(MS2IDdirectory)
annotResult <- annotate(QRYdata = queryFolder, MS2ID = refLibrary)The resulting variable annotResult is an Annot object that contains the annotation results. We can subset its content by applying them getters (custom functions to get the info) or save the whole object (saveRDS function is recommended) for further analysis (more info).
Annotate function offers more features such as prefiltering of the reference spectra library or different similarity metrics. Please, feel free to check them in its own vignette.
Browsing the results
The annotation results can be browsed by using
The
export2xlsxfunction to export the annotation results to an excel fileThe
MS2Iguifunction, which allows to compare graphically both query and reference spectra. Please note that usingMS2IDguifunction with no arguments pops up the interface with no annotation data. Visit the ‘About’ page in MS2IDgui for more information.
MS2IDgui(annotResult)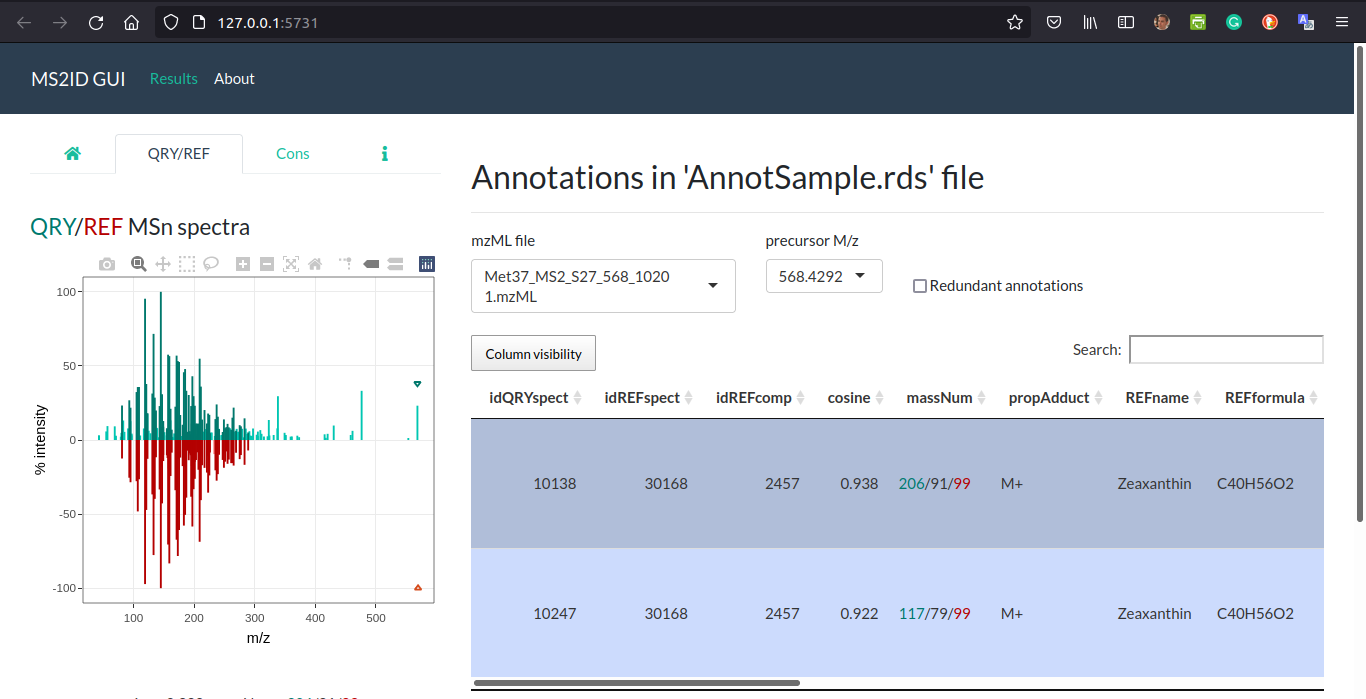
MS2ID GUI interface